ISSN 0253-2778
CN 34-1054/N
It is a fundamental task to ensure the faithful transmission of genetic information across generations for eukaryote species. The centromere is a specialized chromosomal region that is essential for mediating sister chromatid alignment and separation during mitosis. Centromere identity is epigenetically determined by nucleosome-containing centromere protein A (CENP-A). The CENP-A nucleosome provides the foundation for the association of the inner kinetochore and the assembly of the outer kinetochore in mitosis. Here we review centromere identity determination, inner kinetochore function and assembly, and outer kinetochore function and assembly. In particular, we focus on the recent advances in the structure-activity relationship of the constitutive centromere-associated network (CCAN). CCAN structure information sheds new light on our understanding of centromere and kinetochore functions and dynamic organization.

Schematic representation of a mitotic spindle and the organization of core proteins of inner and outer kinetochore.
It is a fundamental task to ensure the faithful transmission of genetic information across generations for eukaryote species. The centromere is a specialized chromosomal region that is essential for mediating sister chromatid alignment and separation during mitosis. Centromere identity is epigenetically determined by nucleosome-containing centromere protein A (CENP-A). The CENP-A nucleosome provides the foundation for the association of the inner kinetochore and the assembly of the outer kinetochore in mitosis. Here we review centromere identity determination, inner kinetochore function and assembly, and outer kinetochore function and assembly. In particular, we focus on the recent advances in the structure-activity relationship of the constitutive centromere-associated network (CCAN). CCAN structure information sheds new light on our understanding of centromere and kinetochore functions and dynamic organization.
The embryonic development and tissue homeostasis maintenance of higher eukaryotes rely on cell proliferation. The cell division cycle (commonly called the cell cycle) is an orderly sequence of cell growth and cell division events that produce two new daughter cells. One cell cycle includes G1 phase, S phase, G2 phase and M phase. The two most important events are DNA synthesis in S phase and sister chromatid alignment and segregation during mitosis. Errors in M phase (including mitosis and cytokinesis) will produce daughter cells with aneuploidy or micronuclei. It is well known that aneuploidy is a hallmark of cancer. Aneuploidy is a type of chromosomal instability and promotes tumorigenesis in a suitable context (e.g., genetic background or microenvironment).
The centromere appears as a constricted chromosome region where the mitotic spindle fibers attach. However, spindle microtubules cannot attach to centromere chromatin directly. Instead, a large protein complex called the kinetochore assembled on the centromere mediates the attachment of spindle microtubules. Nevertheless, the functions of centromeres and kinetochores are highly related and intertwined. In this paper, we summarize the recent advances in centromere function, kinetochore structure and dynamic assembly.
The DNA sequences of centromeres are highly divergent and rapidly evolving[1]. In the budding yeast Saccharomyces cerevisiae, centromeres are defined by a specific ~125-bp DNA sequence, known as a point centromere. Budding yeast kinetochores assemble on point centromeres wrapping a single centromeric nucleosome containing the histone H3 variant Cse4 (the homolog of human CENP-A). Additional Cse4 molecules exist in budding yeast centromeres, but the function and exact molecule number of additional Cse4 is a long-standing debate[2]. Echoing previous studies[3-5], a recent study visualized individual kinetochore complexes in situ in budding yeast using single-molecule localization microscopy[6]. Their finding of 4.8±2.4 copies of Cse4 in a single kinetochore proved the existence of additional noncentromeric copies of Cse4. However, in most eukaryotes, centromeres are not defined by sequence and consist of highly repetitive DNA sequences, such as tandem repeats and retrotransposons, that are unrelated in different organisms. These complex centromeres are known as regional centromeres, and their sizes can extend for several megabases[7, 8]. Regional centromeres vary even between chromosomes of the same organism. Human centromeres are composed of so-called α-satellite repeats, a tandem array of repeat units of approximately 171 bp DNA sequence. Regional centromeres contain multiple CENP-A nucleosomes (CENP-ANuc) and assemble kinetochores that bind multiple microtubules (approximately 20 in humans). In addition to point centromeres and regional centromeres, centromeres extend along the length of entire chromosomes in some species, such as C. elegans, and are called holocentromeres. Holocentromeres have been proposed to be dispersed point centromeres[9].
The sequence and organization of centromeres are not evolutionarily conserved. This fact suggests that the centromere is determined epigenetically[7]. Mounting evidence indicates that CENP-ANuc is the epigenetic marker for centromeres in the majority of eukaryote species[10]. Structural studies indicate that the CATD (CENP-A targeting domain) confers nucleosomal rigidity to CENP-ANuc at the centromere, providing a unique structure that enables the reader to distinguish CENP-A-containing nucleosomes from bulk classical H3 nucleosomes[11, 12].
During S phase, after DNA replication, CENP-A nucleosomes were distributed into two sister chromatids. To maintain centromere stability, new CENP-A molecules must be deposited into centromeres during each cell cycle. Interestingly, new CENP-A molecules are not deposited during S phase or G2 phase in humans. Instead, until cells exit mitosis, new CENP-A molecules deposit into the centromere in G1 phase[13]. However, the cell cycle time windows for CENP-A deposition are not conserved across species. HJURP is the CENP-A chaperone that associates with Mis18α, Mis18β, and Mis18BP1 to target centromeres and mediate the deposition of new CENP-A[14, 15]. During G2 and M phases, higher CDK1 activity phosphorylates both HJURP and Mis18BP1, rendering them inactive and thereafter inhibiting the loading of CENP-A[16, 17]. After mitotic exit, low CDK1 activity licensed the centromere localization of HJURP and CENP-A loading in the next G1 phase.
To separate each sister chromatid accurately into two daughter cells, the mitotic spindle apparatus forms during mitosis. Kinetochores are protein machines linking centromere chromatin with spindle microtubules (Fig. 1). Under an electron microscope, vertebrate kinetochores appear as trilaminar plates, with electron-dense inner and outer kinetochore plates and an electron-lucent middle plate[7]. It is widely accepted to designate the inner plate and outer plate as the inner kinetochore and outer kinetochore, respectively.
First, the inner kinetochore links the centromeric nucleosome directly, and the outer kinetochore attaches spindle microtubules directly. In addition to the physical linker function, kinetochores play multiple functions[18]. A group of proteins localized at the outer kinetochore works as a cell cycle controller through a signaling pathway called the spindle assembly checkpoint (SAC)[19]. Outer kinetochores also provide a platform for enriching several motor proteins, which promote chromosome congression and biorientation. Finally, the chromosome passenger complex, which mainly localizes at the inner centromere but also has a kinetochore proximal pool, plays an error-correction function and is essential for establishing correct kinetochore-microtubule attachment[20].
Inner kinetochores are composed of 16 proteins, which are constitutively localized at the inner kinetochore. Therefore, these proteins are called the constitutive centromere-associated network (CCAN) (Fig. 2). CCAN includes several subcomplexes, CENP-C, CENP-LN, CENP-HIKM, CENP-TWSX and CENP-OPQUR. Among the 16 CCAN proteins, CENP-N and CENP-C bind to CENP-ANuc directly. Straight and colleagues[21] identified CENP-N as a reader of the centromere mark comprising CENP-ANuc. Shortly after the Straight group’s discovery, the Harrison group[22] demonstrated that budding yeast CENP-N forms a complex with CENP-L and determined the structure of the CENP-LN complex. Subsequently, using a combination of genetic, biochemical and cell biology studies, the Cheeseman group[23] demonstrated that the CENP-LN complex is critical for CCAN assembly. Their work also demonstrated that multiple interactions among different CCAN subcomplexes determine each subcomplex’s localization and CCAN integrity.
In addition to CENP-N, CENP-C is another CCAN component that directly interacts with CENP-ANuc[24]. In vitro arrays of CENP-ANuc recruit CENP-C via the CENP-A C-terminal LEEGLG motif[25]. A structural comparison of the human CENP-A and H3 nucleosomes revealed that CENP-A contains two extra amino acid residues (Arg 80 and Gly 81) in the RG-loop (also called the L1 loop)[26]. Indeed, Fang et al.[27] demonstrated that the RG-loop of CENP-A is required for recruiting CENP-N to centromeres and faithful mitosis. Later, three groups independently reported the structure of CENP-N binding to CENP-ANuc[28-30]. These structural, biochemical and cell biology studies confirmed that CENP-N is a reader for CENP-ANuc and verified the importance of the RG-loop in mediating CENP-N binding. In addition, the CENP-N-CENP-ANuc interaction is stabilized by electrostatic interactions between CENP-N and nucleosomal DNA.
The structure of an intact CCAN bound to CENP-ANuc is a “holy grail” for structural biologists and cell biologists who study mitosis and chromosomes. The Barford group[31] uncovered the cryo-electron microscopy structure of the Saccharomyces cerevisiae CCAN complex assembled onto a CENP-ANuc (CCAN-CENP-ANuc). The Y-shaped overall CCAN structure explains the interdependency of the different subcomplexes of CCAN and shows how the opening of CCAN accommodates CENP-ANuc. Importantly, CENP-LN interacts with the unwrapped DNA duplex at the two termini of CENP-ANuc. Biochemical and genetic studies verified that the CENP-N DNA-binding groove is required for stable CCAN-CENP-ANuc interaction.
Mounting evidence has demonstrated that CCAN components are largely conserved between fungi and mammals. Recently, the Barford group, Musacchio group and our group independently uncovered the structure of human CCAN[32-34]. Indeed, the overall architecture of human CCAN is similar to that of budding yeast CCAN. To solve the structure of CCAN-CENP-ANuc, we reconstituted a 16-subunit human CCAN complex assembled on CENP-ANuc wrapped with 147 bp of DNA using the Widom 601 sequence. We note that Widom 601 DNA has high affinity for histone octamers, but its DNA sequence is not the natural centromere sequence. In the human CCAN structure we solved, there are four subcomplexes, CENP-LN, CENP-HIKM, CENP-TWSX, and CENP-OPQUR. The arrangement of the four subcomplexes generates a b-shaped structure, in which CENP-OPQUR adopts an elongated shape to generate the arm, and CENP-LN, CENP-HIKM, and CENP-TWSX form the semicircle. The CENP-LN subcomplex, located at the center of the ‘b’, functions as a node for coordination of the assembly of CCAN by contributing the contact sites, with the subcomplexes CENP-HIKM and CENP-OPQU on the opposite side[32]. In general, the human CCAN structure reported by two other groups is consistent with our data.
To our surprise, a DNA double helix approximately 25 bp in length was clearly resolved in our CCAN structure, although the CENP-ANuc signal could not be determined. Around the DNA is a positively charged channel composed of CENP-LN, CENP-HIKhead, and CENP-TW, which complements the negative charge of the DNA gyre. As shown in Fig. 3a, the CCAN complex binds to double-stranded DNA through electrostatic interactions between a set of positively charged residues from several CCAN components and the negatively charged phosphate backbone of DNA[32]. In agreement with our observation, Barford’s structure also concludes that the positively charged CCAN channel grips the linker α-satellite DNA of CENP-ANuc. Through biochemical and cell biology studies, we further confirmed the importance of key positively charged residues of CENP-LN in the assembly of CCAN and faithful chromosome segregation during mitosis. For instance, CENP-L mutants with mutations in the four positively charged residues failed to localize to the kinetochore effectively (Fig. 3b, c).
In yeast, the Y-shape of CCAN opening can accommodate CENP-ANuc to enable specific CCAN subunits to contact the nucleosomal DNA and histone subunits[31]. In contrast, human CCAN forms edge-on contacts with CENP-ANuc, and the CENP-LN channel grips the linker DNA of CENP-ANuc[32, 33]. Topological entrapment of the linker DNA by CCAN likely provides a molecular explanation for the tight kinetochore assembly on the centromere to withstand the pulling force during chromosome movement and segregation.
The main function of CCAN is linking centromere CENP-ANuc with the outer kinetochore. Although CCAN proteins bind centromeres throughout the cell cycle, these proteins are phosphorylated during mitosis and have more functions. Considering its multiple binding activity with CENP-A, CENP-LN and CENP-HIKM, CENP-C is generally recognized as a blueprint for directing CCAN assembly. CENP-C has additional role in recruiting M18BP1 to centromeres to promote CENP-A chromatin assembly[35]. CDK1 phosphorylates CENP-C and boosts the binding between CENP-C and CENP-ANuc[36]. We note that the CENP-C-CENP-ANuc interaction is important since CENP-ANuc does not directly bind to other CCAN components in mitosis. The CENP-LN subcomplex forms a channel to bind the linker DNA of CENP-ANuc in both budding yeast and humans. Interestingly, both CENP-L and CENP-N are substrates of CDK1. Our study demonstrated that phosphorylation of CENP-N Ser299 by CDK1 disrupts the CENP-N/L interaction and CENP-N kinetochore localization[37]. Consistent with our work, a study from the Cheeseman group[38] also concluded that phosphorylation of CENP-L and CENP-N controls CENP-LN complex formation and localization in a cell cycle-dependent manner.
CENP-U functions as a receptor to recruit PLK1 to the kinetochore, stabilizing kinetochore-microtubule attachment[39, 40]. Our group recently demonstrated that phosphorylation of CENP-R by Aurora B regulates kinetochore-microtubule attachment for accurate chromosome segregation[41]. We summarized the reported CDK1 phosphorylation sites toward CCAN components in Table 1.
| Substrates | Sites | Function | References |
| CENP-C | T651 (chicken)
T734 (human) |
CDK1-mediated phosphorylation of CENP-C facilitates its binding to CENP-A in vitro and in vivo. | [36, 42] |
| CENP-T | T11,
T85, S201, Multiple sites |
CDK1 phosphorylation towards CENP-T promote kinetochore assembly during mitosis. Phosphorylation of CENP-T at Thr11 or Thr85 is sufficient to recruit Ndc80 complex. Phosphorylated Ser201 of CENP-T is capable to bind the Mis12 complex at low CDK1 activity. | [51, 53, 57] |
| CENP-U | T98 (human) | CDK1 and PLK1 phosphorylation towards CENP-U enable the kinetochore recruitment of PLK1 by CENP-U. | [39] |
| T31, S41, S45, S53 (budding yeast) | Cdk1 phosphorylation activates phospho-degrons on the essential subunit Ame1/CENP-U. | [43] | |
| CENP-N | T158, T172, T220, S299, S320 | Phosphorylation of CENP-N by CDK1 during mitosis negatively regulates its kinetochore localization, and dynamic phosphorylation plays an important role in the precise function of CENP-LN. | [37, 38] |
| CENP-L | T10, T25, T43, T78, S209, S246, T271 | Phosphorylation of CENP-L by CDK1 during mitosis negatively regulates its kinetochore localization, and dynamic phosphorylation plays an important role in the precise function of CENP-LN. | [38] |
The core outer kinetochore proteins comprise a conserved network called KMN, including the Knl1-Zwint1 subcomplex, the Mis12 complex (Mis12C, which contains Mis12, Dsn1/Mis13, Nsl1/Mis14 and Nnf1) and the Ndc80 complex (Ndc80C, which contains Hec1/Ndc80, Nuf2, Spc24 and Spc25)[44, 45]. In contrast to CCAN proteins, which constitutively bind centromeres, the KMN network and other outer kinetochore proteins are rapidly assembled on the inner kinetochore when cells enter mitosis. Recent work has highlighted two parallel pathways for the assembly of the KMN network (Fig. 2). The N-terminal region of CENP-C recruits Mis12C through direct binding between the N-terminal region of CENP-C and Mis12C[46, 47]. In turn, Mis12C can recruit Knl1 (in complex with Zwint1) and Ndc80C[48]. The other pathway depends on the CENP-TWSX subcomplex, which can bind centromeric DNA. CENP-T can subsequently recruit Ndc80C via direct binding between the CENP-T N-terminal tail and the Spc24/Spc25 subunit of Ndc80C upon mitotic entry[49, 50]. In summary, CENP-C and CENP-T mediate two parallel pathways for outer kinetochore assembly.
Several mechanisms determine the timing of KMN assembly in prophase. The Ndc80C is sequestered outside the nucleus throughout interphase and is thereby spatially separated from the CCAN until mitosis[51]. KMN assembly is promoted by CDK1 and Aurora B, whose kinase activity peaks in mitosis. CDK1 phosphorylation of CENP-T promotes the direct interaction of CENP-T with Ndc80C and Mis12C[52, 53]. In addition, the phosphorylation of Dsn1/Mis13 by Aurora B enhances the interaction between CENP-C and Mis12C during mitosis[48, 54, 55]. Interestingly, phosphorylation of fission yeast CENP-C by Aurora B impairs the CENP-C-Mis12C interaction[56]. Another study from the Fukagawa group investigated the contribution of the CENP-C and CENP-T pathways in the recruitment of the KMN network to kinetochores and the phosphorylation regulation of these two pathways during mitotic progression[57]. These studies suggest the existence of remarkable phosphorylation-regulated plasticity in the inner-outer kinetochore interface during different mitotic stages and in different species/cells.
The KMN network then scaffolds the localization of other outer kinetochore proteins, such as proteins involved in the spindle assembly checkpoint (SAC) and mitotic motor proteins. Here, we briefly summarize the recruitment of core SAC proteins. Ndc80C is the major kinetochore site for microtubule binding (through its Hec1 and Nuf2 subunits)[58]. Interestingly, the kinetochore localization of Mps1 kinase, an upstream kinase of SAC signaling, also relies on Hec1 and Nuf2[59, 60]. Thus, Mps1 and microtubules competitively bind Ndc80C, which provides a wonderful molecular mechanism for how the kinetochore couples microtubule attachment with SAC signaling[61, 62]. The Mps1 phosphorylation of Knl1 on conserved MELT motifs initiates the recruitment of Bub3/Bub1 and Bub3/BubR1 since Bub3 functions as a reader for phosphorylated MELpT[63]. In turn, Bub1 subsequently recruits Mad1/Mad2. Intriguingly, Mps1 activity is essential for Bub1’s recruitment of Mad1/Mad2[64]. As the key component of the mitotic checkpoint complex, the recruitment of Mad1/Mad2 also depends on the RZZ complex (Rod, ZW10, Zwilch)[65].
Cyclin-dependent kinase 1 (CDK1) is a master kinase that coordinates mitotic progression and requires its regulatory subunit Cyclin B to ensure full kinase activity and substrate specificity. Both Cyclin B1 and B2 localize at the outer kinetochores during mitosis. Barr and colleagues[66] have established the Cyclin B1-Mad1 interaction and have revealed the role of Cyclin B1 as a bona fide SAC component. A study by the Saurin group[67] showed that Cyclin B1 scaffolds the kinetochore corona localization of Mad1 and ensures a robust SAC signal. Furthermore, a study from Pines’ group[68] demonstrated that Mad1 recruits Cyclin B1-Cdk1 to nuclear pores. In turn, Cyclin B1-Cdk1 promotes Mad1 translocation to kinetochores from nuclear pores. Despite some inconsistencies, these three independent studies demonstrated the importance of Cyclin B1 kinetochore localization in robust SAC signaling. Interestingly, our recent study demonstrated that Cyclin B2 selectively localizes at the unattached kinetochores in prometaphase and established that Mad2 promotes Cyclin B2 recruitment to the kinetochore for faithful mitotic progression[69].
Combinations of advanced optical imaging protocols, such as lattice light-sheet microscopy with adaptive optics and photoactivatable complementary fluorescence, spectral imaging analyses, and correlative light and cryo-electron microscopic tomography, would increase our understanding of CCAN assembly dynamics during the cell cycle and CCAN communication with the KMN network[70, 71]. The newly developed lattice light-sheet microscopy with adaptive optics has enabled, for live organoids, noninvasive, aberration-free imaging of subcellular processes, including kinetochore-microtubule attachment switching from lateral attachment to end-on capture during mitosis[72]. In this context, inclusion of the expanding collection of gene-edited organoids will allow modeling of chromosome interactions and CCAN regulation underlying the control of cell division[73].
The large size and complex architecture of the centromere, which contains numerous proteins possessing low-complexity regions, are linked to its regulatory mechanisms involving posttranslational modifications and interchangeable complex subunits between compartments in mitosis. Thus, the centromere must possess intrinsic self-control mechanisms. According to proteomic and bioinformatics analyses, the centromere is composed of perhaps 15 scaffolding proteins with a total of more than 150 proteins[70]. Future work will describe the spatiotemporal dynamics and physicochemical properties of these low-complexity kinetochore proteins driven by LLPS during cell division, as recently reported[74–76]. Finally, recent clinical and translational studies support our early rationale that genetic variation affects protein posttranslational modifications and is involved in rewiring biological pathways to generate asymmetric division of tumor cells.
The CCAN structure provides new insight into centromere and kinetochore organization and a foundation for developing chemical tools to delineate the spatiotemporal dynamics of centromere assembly and communication. The substantial overall architecture similarity between budding yeast CCAN (scCCAN) and human CCAN (hsCCAN) suggests a common evolutionary origin of centromere kinetochore assembly. Comparison of the structure of CENP-N-CENP-ANuc with that of human CCAN-CENP-ANuc suggests that there is a large conformational change or allosteric effect when the whole CCAN is assembled. Cryo-EM can capture a wide variety of conformational states of macromolecules in solution. Conformational changes are often transient but can be trapped by vitrification at specific time points following the initiation of the reaction and imaged using time-resolved cryo-EM[77]. It would be of great interest and excitement to visualize the CENP-N conformational change during CCAN assembly using time-resolved cryo-EM and to address the underlying mechanism.
Despite progress over the past decade in understanding the function and mechanism of centromere assembly and plasticity control, much remains to be explored. We still have a minimal understanding of the molecular mechanisms underlying centromere assembly/disassembly during the cell cycle. Advances will require cryo-electron tomographic analyses of the different states of the centromeres of mitotic cells in situ. Currently, it is unclear to what extent CCAN structural perturbation results in chromothripsis[78]. Thus, the challenges ahead are to delineate and distinguish the characteristics of chromosome-specific CCAN assembly that exhibit susceptibility to chromosome segregation effort and CCAN assembly. Overall, we anticipate that advances in our understanding of the molecular language of centromere network communications will enable us to consolidate temporal protein interactions into a working model for decision making in cell division and targeted interrogation for aberrant CCAN-elicited pathogenesis[79].
We are grateful to Prof. Yunyu Shi for her support. We thank members of our laboratory for insightful discussion. This work was supported by the National Key R&D Program of China (2022YFA1303100, 2022YFA0806800), the National Natural Science Foundation of China (32090040, 31621002, 31871359, 32170733, 92153302), the Ministry of Education (IRT_17R102, 20113402130010, YD2070006001), and the Fundamental Research Funds for the Central Universities (WK2070000194).
The authors declare that they have no conflict of interest.
| [1] |
McAinsh A D, Marston A L. The four causes: The functional architecture of centromeres and kinetochores. Annu. Rev. Genet., 2022, 56: 279–314. DOI: 10.1146/annurev-genet-072820-034559
|
| [2] |
Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics, 2013, 194 (4): 817–846. DOI: 10.1534/genetics.112.145276
|
| [3] |
Lawrimore J, Bloom K S, Salmon E D. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol., 2011, 195 (4): 573–582. DOI: 10.1083/jcb.201106036
|
| [4] |
Coffman V C, Wu P C, Parthun M R, et al. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol., 2011, 195 (4): 563–572. DOI: 10.1083/jcb.201106078
|
| [5] |
Haase J, Mishra P K, Stephens A, et al. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr. Biol., 2013, 23 (19): 1939–1944. DOI: 10.1016/j.cub.2013.07.083
|
| [6] |
Cieslinski K, Wu Y L, Nechyporenko L, et al. Nanoscale structural organization and stoichiometry of the budding yeast kinetochore. J. Cell Biol., 2023, 222 (4): e202209094. DOI: 10.1083/jcb.202209094
|
| [7] |
Cleveland D W, Mao Y, Sullivan K F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell, 2003, 112 (4): 407–421. DOI: 10.1016/S0092-8674(03)00115-6
|
| [8] |
Allshire R C, Karpen G H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet., 2008, 9 (12): 923–937. DOI: 10.1038/nrg2466
|
| [9] |
Steiner F A, Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife, 2014, 3: e02025. DOI: 10.7554/eLife.02025
|
| [10] |
Kixmoeller K, Allu P K, Black B E. The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol., 2020, 10 (6): 200051. DOI: 10.1098/rsob.200051
|
| [11] |
Black B E, Foltz D R, Chakravarthy S, et al. Structural determinants for generating centromeric chromatin. Nature, 2004, 430 (6999): 578–582. DOI: 10.1038/nature02766
|
| [12] |
Sekulic N, Bassett E A, Rogers D J, et al. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature, 2010, 467 (7313): 347–351. DOI: 10.1038/nature09323
|
| [13] |
Jansen L E T, Black B E, Foltz D R, et al. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol., 2007, 176 (6): 795–805. DOI: 10.1083/jcb.200701066
|
| [14] |
Foltz D R, Jansen L E T, Bailey A O, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell, 2009, 137 (3): 472–484. DOI: 10.1016/j.cell.2009.02.039
|
| [15] |
Dunleavy E M, Roche D, Tagami H, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell, 2009, 137 (3): 485–497. DOI: 10.1016/j.cell.2009.02.040
|
| [16] |
Wang J Y, Liu X, Dou Z, et al. Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP). J. Biol. Chem., 2014, 289 (12): 8326–8336. DOI: 10.1074/jbc.M113.529958
|
| [17] |
Stankovic A, Guo L Y, Mata J F, et al. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol. Cell, 2017, 65 (2): 231–246. DOI: 10.1016/j.molcel.2016.11.021
|
| [18] |
Pesenti M E, Weir J R, Musacchio A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol., 2016, 37: 152–163. DOI: 10.1016/j.sbi.2016.03.003
|
| [19] |
Dou Z, Prifti D K, Gui P, et al. Recent progress on the localization of the spindle assembly checkpoint machinery to kinetochores. Cells, 2019, 8 (3): 278. DOI: 10.3390/cells8030278
|
| [20] |
Carmena M, Wheelock M, Funabiki H, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol., 2012, 13 (12): 789–803. DOI: 10.1038/nrm3474
|
| [21] |
Carroll C W, Silva M C C, Godek K M, et al. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol., 2009, 11 (7): 896–902. DOI: 10.1038/ncb1899
|
| [22] |
Hinshaw S M, Harrison S C. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep., 2013, 5 (1): 29–36. DOI: 10.1016/j.celrep.2013.08.036
|
| [23] |
McKinley K L, Sekulic N, Guo L Y, et al. The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell, 2015, 60 (6): 886–898. DOI: 10.1016/j.molcel.2015.10.027
|
| [24] |
Carroll C W, Milks K J, Straight A F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol., 2010, 189 (7): 1143–1155. DOI: 10.1083/jcb.201001013
|
| [25] |
Guse A, Carroll C W, Moree B, et al. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature, 2011, 477 (7364): 354–358. DOI: 10.1038/nature10379
|
| [26] |
Tachiwana H, Kagawa W, Shiga T, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature, 2011, 476 (7359): 232–235. DOI: 10.1038/nature10258
|
| [27] |
Fang J N, Liu Y T, Wei Y, et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev., 2015, 29 (10): 1058–1073. DOI: 10.1101/gad.259432.115
|
| [28] |
Tian T, Li X R, Liu Y Y, et al. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res., 2018, 28 (3): 374–378. DOI: 10.1038/cr.2018.13
|
| [29] |
Chittori S, Hong J, Saunders H, et al. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science, 2018, 359 (6373): 339–343. DOI: 10.1126/science.aar2781
|
| [30] |
Pentakota S, Zhou K, Smith C, et al. Decoding the centromeric nucleosome through CENP-N. eLife, 2017, 6: e33442. DOI: 10.7554/eLife.33442
|
| [31] |
Yan K, Yang J, Zhang Z, et al. Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature, 2019, 574 (7777): 278–282. DOI: 10.1038/s41586-019-1609-1
|
| [32] |
Tian T, Chen L, Dou Z, et al. Structural insights into human CCAN complex assembled onto DNA. Cell Discov., 2022, 8 (1): 90. DOI: 10.1038/s41421-022-00439-6
|
| [33] |
Yatskevich S, Muir K W, Bellini D, et al. Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome. Science, 2022, 376 (6595): 844–852. DOI: 10.1126/science.abn3810
|
| [34] |
Pesenti M E, Raisch T, Conti D, et al. Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization. Mol. Cell, 2022, 82 (11): 2113–2131.e8. DOI: 10.1016/j.molcel.2022.04.027
|
| [35] |
Moree B, Meyer C B, Fuller C J, et al. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol., 2011, 194 (6): 855–871. DOI: 10.1083/jcb.201106079
|
| [36] |
Watanabe R, Hara M, Okumura E I, et al. CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J. Cell Biol., 2019, 218 (218): 4042–4062. DOI: 10.1083/jcb.201907006
|
| [37] |
Liu, R, Dou, Z, Tian, T, et al. Dynamic phosphorylation of CENP-N by CDK1 guides accurate chromosome segregation in mitosis. J. Mol. Cell Biol., 2023: mjad041. DOI: 10.1093/jmcb/mjad041
|
| [38] |
Navarro A P, Cheeseman I M. Dynamic cell cycle-dependent phosphorylation modulates CENP-L-CENP-N centromere recruitment. Mol. Biol. Cell, 2022, 33 (10): ar87. DOI: 10.1091/mbc.E22-06-0239
|
| [39] |
Singh P, Pesenti M E, Maffini S, et al. BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore receptors in mitosis. Mol. Cell, 2021, 81 (1): 67–87.e9. DOI: 10.1016/j.molcel.2020.10.040
|
| [40] |
Chen Q F, Zhang M, Pan X, et al. Bub1 and CENP-U redundantly recruit Plk1 to stabilize kinetochore-microtubule attachments and ensure accurate chromosome segregation. Cell Rep., 2021, 36 (12): 109740. DOI: 10.1016/j.celrep.2021.109740
|
| [41] |
Sedzro D M, Yuan X, Mullen M, et al. Phosphorylation of CENP-R by Aurora B regulates kinetochore-microtubule attachment for accurate chromosome segregation. J. Mol. Cell Biol., 2022, 14 (7): mjac051. DOI: 10.1093/jmcb/mjac051
|
| [42] |
Ariyoshi M, Makino F, Watanabe R, et al. Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C. EMBO J., 2021, 40 (5): e105671. DOI: 10.15252/embj.2020105671
|
| [43] |
Böhm M, Killinger K, Dudziak A, et al. Cdc4 phospho-degrons allow differential regulation of Ame1CENP-U protein stability across the cell cycle. eLife, 2021, 10: e67390. DOI: 10.7554/eLife.67390
|
| [44] |
Westermann S, Cheeseman I M, Anderson S, et al. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol., 2003, 163 (2): 215–222. DOI: 10.1083/jcb.200305100
|
| [45] |
Cheeseman I M, Niessen S, Anderson S, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev., 2004, 18 (18): 2255–2268. DOI: 10.1101/gad.1234104
|
| [46] |
Screpanti E, De Antoni A, Alushin G M, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol., 2011, 21 (5): 391–398. DOI: 10.1016/j.cub.2010.12.039
|
| [47] |
Przewloka M R, Venkei Z, Bolanos-Garcia V M, et al. CENP-C is a structural platform for kinetochore assembly. Curr. Biol., 2011, 21 (5): 399–405. DOI: 10.1016/j.cub.2011.02.005
|
| [48] |
Petrovic A, Keller J, Liu Y, et al. Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell, 2016, 167 (4): 1028–1040.e15. DOI: 10.1016/j.cell.2016.10.005
|
| [49] |
Gascoigne K E, Takeuchi K, Suzuki A, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell, 2011, 145 (3): 410–422. DOI: 10.1016/j.cell.2011.03.031
|
| [50] |
Schleiffer A, Maier M, Litos G, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol., 2012, 14 (6): 604–613. DOI: 10.1038/ncb2493
|
| [51] |
Gascoigne K E, Cheeseman I M. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol., 2013, 201 (1): 23–32. DOI: 10.1083/jcb.201301006
|
| [52] |
Nishino T, Rago F, Hori T, et al. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J., 2013, 32 (3): 424–436. DOI: 10.1038/emboj.2012.348
|
| [53] |
Huis In 't Veld P J, Jeganathan S, Petrovic A, et al. Molecular basis of outer kinetochore assembly on CENP-T. eLife, 2016, 5: e21007. DOI: 10.7554/eLife.21007
|
| [54] |
Yang Y, Wu F, Ward T, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem., 2008, 283 (39): 26726–26736. DOI: 10.1074/jbc.M804207200
|
| [55] |
Dimitrova Y N, Jenni S, Valverde R, et al. Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell, 2016, 167 (4): 1014–1027.e12. DOI: 10.1016/j.cell.2016.10.011
|
| [56] |
Zhou X, Zheng F, Wang C L, et al. Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis. Proc. Natl. Acad. Sci. U.S.A., 2017, 114 (50): E10667–E10676. DOI: 10.1073/pnas.1710506114
|
| [57] |
Hara M, Ariyoshi M, Okumura E I, et al. Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat. Cell Biol., 2018, 20 (12): 1378–1388. DOI: 10.1038/s41556-018-0230-0
|
| [58] |
Foley E A, Kapoor T M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol., 2013, 14 (1): 25–37. DOI: 10.1038/nrm3494
|
| [59] |
Nijenhuis W, von Castelmur E, Littler D, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol., 2013, 201 (2): 217–231. DOI: 10.1083/jcb.201210033
|
| [60] |
Dou Z, Liu X, Wang W W, et al. Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl. Acad. Sci. U.S.A., 2015, 112 (33): E4546–E4555. DOI: 10.1073/pnas.1508791112
|
| [61] |
Hiruma Y, Sacristan C, Pachis S T, et al. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science, 2015, 348 (6240): 1264–1267. DOI: 10.1126/science.aaa4055
|
| [62] |
Ji Z, Gao H, Yu H. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science, 2015, 348 (6240): 1260–1264. DOI: 10.1126/science.aaa4029
|
| [63] |
Primorac I, Weir J R, Chiroli E, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife, 2013, 2: e01030. DOI: 10.7554/eLife.01030
|
| [64] |
Ji Z, Gao H, Jia L, et al. A sequential multitarget Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife, 2017, 6: e22513. DOI: 10.7554/eLife.22513
|
| [65] |
Rodriguez-Rodriguez J A, Lewis C, McKinley K L, et al. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol., 2018, 28 (21): 3422–3429.e5. DOI: 10.1016/j.cub.2018.10.006
|
| [66] |
Alfonso-Pérez T, Hayward D, Holder J, et al. MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J. Cell Biol., 2019, 218 (4): 1108–1117. DOI: 10.1083/jcb.201808015
|
| [67] |
Allan L A, Reis M C, Ciossani G, et al. Cyclin B1 scaffolds MAD1 at the kinetochore corona to activate the mitotic checkpoint. EMBO J., 2020, 39 (12): e103180. DOI: 10.15252/embj.2019103180
|
| [68] |
Jackman M, Marcozzi C, Barbiero M, et al. Cyclin B1-Cdk1 facilitates MAD1 release from the nuclear pore to ensure a robust spindle checkpoint. J. Cell Biol., 2020, 219 (6): e201907082. DOI: 10.1083/jcb.201907082
|
| [69] |
Liu S K, Yuan X, Cui P, et al. Mad2 promotes Cyclin B2 recruitment to the kinetochore for guiding accurate mitotic checkpoint. EMBO Rep., 2022, 23 (6): e54171. DOI: 10.15252/embr.202154171
|
| [70] |
Liu X, Liu X, Wang H W, et al. Phase separation drives decision making in cell division. J Biol. Chem., 2020, 295 (39): 13419–13431. DOI: 10.1074/jbc.REV120.011746
|
| [71] |
Xia P, Liu X, Wu B, et al. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell, 2014, 25 (25): 4166–4173. DOI: 10.1091/mbc.E14-06-1133
|
| [72] |
Huang Y J, Lin L, Liu X, et al. BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res., 2019, 29 (7): 562–578. DOI: 10.1038/s41422-019-0178-z
|
| [73] |
Song X Y, Yang F R, Liu X, et al. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat. Chem. Biol., 2021, 17 (12): 1314–1323. DOI: 10.1038/s41589-021-00875-7
|
| [74] |
Song X Y, Yang F R, Yang T T, et al. Phase separation of EB1 guides microtubule plus-end dynamics. Nat. Cell Biol., 2023, 25 (1): 79–91. DOI: 10.1038/s41556-022-01033-4
|
| [75] |
Meier S M, Farcas A M, Kumar A, et al. Multivalency ensures persistence of a +TIP body at specialized microtubule ends. Nat. Cell Biol., 2023, 25 (1): 56–67. DOI: 10.1038/s41556-022-01035-2
|
| [76] |
Maan R, Reese L, Volkov V A, et al. Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus-ends. Nat. Cell Biol., 2023, 25 (1): 68–78. DOI: 10.1038/s41556-022-01037-0
|
| [77] |
Zhang S W, Zou S T, Yin D Y, et al. USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Nature, 2022, 605 (7910): 567–574. DOI: 10.1038/s41586-022-04671-8
|
| [78] |
Ly P, Brunner S F, Shoshani O, et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet., 2019, 51 (4): 705–715. DOI: 10.1038/s41588-019-0360-8
|
| [79] |
Yao X B, Smolka A J. Gastric parietal cell physiology and Helicobacter pylori-induced disease. Gastroenterology, 2019, 156 (8): 2158–2173. DOI: 10.1053/j.gastro.2019.02.036
|
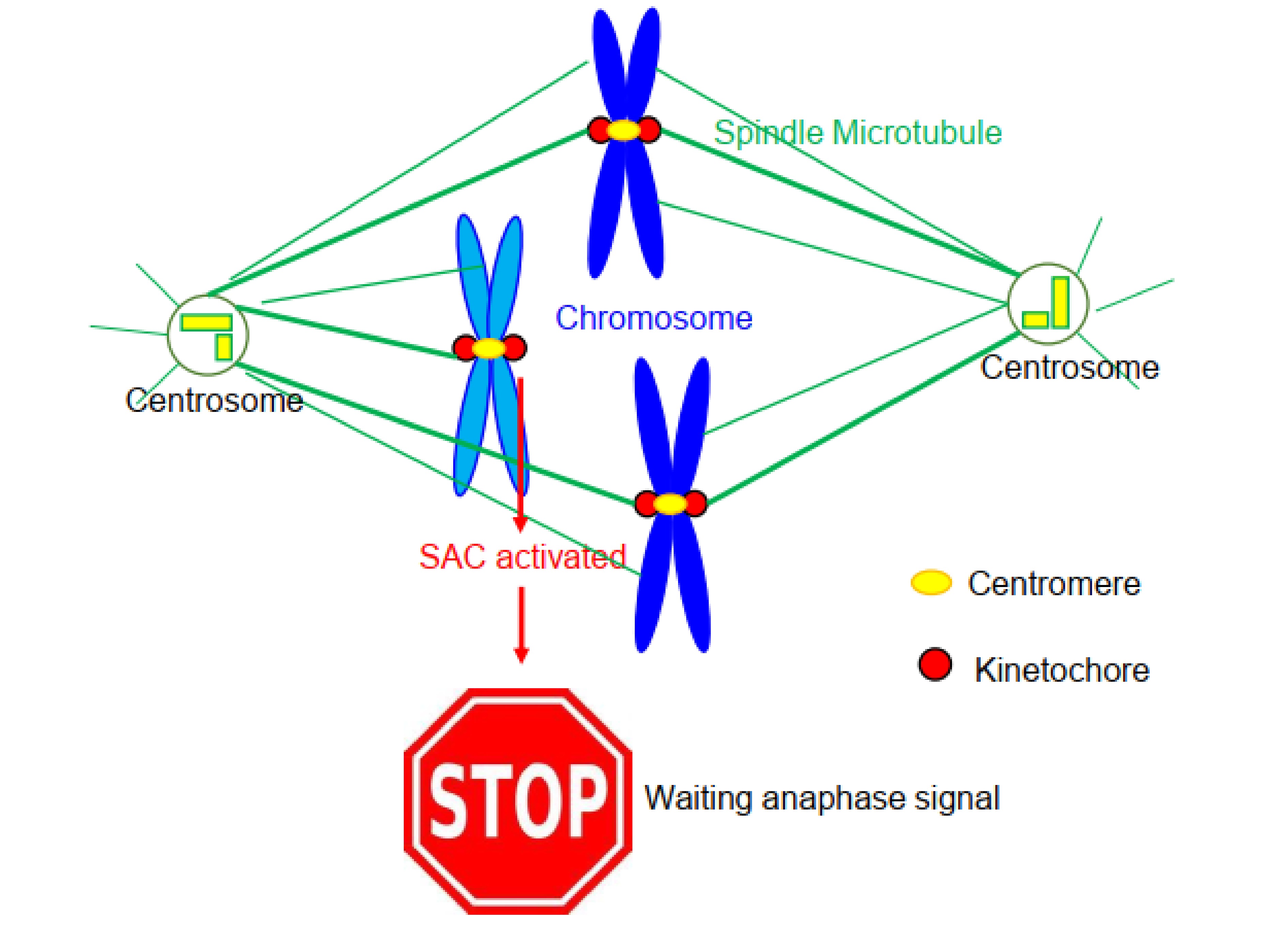
Figure 1. A cartoon shown mitotic spindle and the chromosomes. In the central position of the spindle, two chromosomes achieved correct microtubule attachment. On the contrary, a chromosome near the left pole failed to establish proper microtubule attachment. Thus, the kinetochores of this unaligned chromosome initiate a signaling call the spindle assembly checkpoint (SAC) and thereafter preclude the cell to enter anaphase. The yellow dots on chromosomes depict centromeres, and the red dots depict kinetochores.
Figure 2. The structure of inner kinetochore and outer kinetochore core proteins. Centromeric CENP-A nucleosome serves as the foundation for kinetochore assembly. CCAN, a complex comprising 16 proteins, constitutively localizes at the centromeres. The cartoon shows the 3D architecture of CCAN bound CENP-A nucleosome. Interestingly, CCAN forms a channel that topologically grasps linker DNA of CENP-A nucleosome. CENP-T and CENP-C, which both have an elongated N-terminal tail, functions as two parallel pathways to recruit Knl1 complex, Mis12 complex and Ndc80 complex in mitosis. Knl1 complex, Mis12 complex and Ndc80 complex comprise the core of outer kinetochore and were named KMN network. KMN harbors two activity, attaching the spindle microtubule and recruiting SAC signaling proteins.
Figure 3. DNA binds to CCAN through the CENP-LN channel[32]. (a) Electrostatic potential surface view of CENP-LN-HIK head-TW binding with DNA. The DNA is shown as cartoon. Note that positively charged amino acids from CENPLN, CENP-I and CENP-TW constitute the contact sites between CCAN and DNA. (b) Representative immunofluorescence montage of HeLa cells expressing GFPCENP-L wild type and DNA binding-deficient mutants. 4KA represents CENP-L K155A/R306A/K319A/K321A, 4KE represents K155E/R306E/K319E/K321E. Scale bar, 10 μm. (c) Statistical analysis of kinetochore intensity of various GFP-CENP-L mutants as treated in b. Bars represent the mean kinetochore intensity (±SEM) normalized to values of the CENP-L WT expressing group. Each dot represents one cell (30 cells from three independent experiments). Ordinary one-way ANOVA followed by Tukey’s post hoc test was used to determine statistical significance. ****p < 0.0001.
| [1] |
McAinsh A D, Marston A L. The four causes: The functional architecture of centromeres and kinetochores. Annu. Rev. Genet., 2022, 56: 279–314. DOI: 10.1146/annurev-genet-072820-034559
|
| [2] |
Biggins S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics, 2013, 194 (4): 817–846. DOI: 10.1534/genetics.112.145276
|
| [3] |
Lawrimore J, Bloom K S, Salmon E D. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J. Cell Biol., 2011, 195 (4): 573–582. DOI: 10.1083/jcb.201106036
|
| [4] |
Coffman V C, Wu P C, Parthun M R, et al. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J. Cell Biol., 2011, 195 (4): 563–572. DOI: 10.1083/jcb.201106078
|
| [5] |
Haase J, Mishra P K, Stephens A, et al. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr. Biol., 2013, 23 (19): 1939–1944. DOI: 10.1016/j.cub.2013.07.083
|
| [6] |
Cieslinski K, Wu Y L, Nechyporenko L, et al. Nanoscale structural organization and stoichiometry of the budding yeast kinetochore. J. Cell Biol., 2023, 222 (4): e202209094. DOI: 10.1083/jcb.202209094
|
| [7] |
Cleveland D W, Mao Y, Sullivan K F. Centromeres and kinetochores: From epigenetics to mitotic checkpoint signaling. Cell, 2003, 112 (4): 407–421. DOI: 10.1016/S0092-8674(03)00115-6
|
| [8] |
Allshire R C, Karpen G H. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat. Rev. Genet., 2008, 9 (12): 923–937. DOI: 10.1038/nrg2466
|
| [9] |
Steiner F A, Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife, 2014, 3: e02025. DOI: 10.7554/eLife.02025
|
| [10] |
Kixmoeller K, Allu P K, Black B E. The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol., 2020, 10 (6): 200051. DOI: 10.1098/rsob.200051
|
| [11] |
Black B E, Foltz D R, Chakravarthy S, et al. Structural determinants for generating centromeric chromatin. Nature, 2004, 430 (6999): 578–582. DOI: 10.1038/nature02766
|
| [12] |
Sekulic N, Bassett E A, Rogers D J, et al. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature, 2010, 467 (7313): 347–351. DOI: 10.1038/nature09323
|
| [13] |
Jansen L E T, Black B E, Foltz D R, et al. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol., 2007, 176 (6): 795–805. DOI: 10.1083/jcb.200701066
|
| [14] |
Foltz D R, Jansen L E T, Bailey A O, et al. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell, 2009, 137 (3): 472–484. DOI: 10.1016/j.cell.2009.02.039
|
| [15] |
Dunleavy E M, Roche D, Tagami H, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell, 2009, 137 (3): 485–497. DOI: 10.1016/j.cell.2009.02.040
|
| [16] |
Wang J Y, Liu X, Dou Z, et al. Mitotic regulator Mis18β interacts with and specifies the centromeric assembly of molecular chaperone holliday junction recognition protein (HJURP). J. Biol. Chem., 2014, 289 (12): 8326–8336. DOI: 10.1074/jbc.M113.529958
|
| [17] |
Stankovic A, Guo L Y, Mata J F, et al. A dual inhibitory mechanism sufficient to maintain cell-cycle-restricted CENP-A assembly. Mol. Cell, 2017, 65 (2): 231–246. DOI: 10.1016/j.molcel.2016.11.021
|
| [18] |
Pesenti M E, Weir J R, Musacchio A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol., 2016, 37: 152–163. DOI: 10.1016/j.sbi.2016.03.003
|
| [19] |
Dou Z, Prifti D K, Gui P, et al. Recent progress on the localization of the spindle assembly checkpoint machinery to kinetochores. Cells, 2019, 8 (3): 278. DOI: 10.3390/cells8030278
|
| [20] |
Carmena M, Wheelock M, Funabiki H, et al. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol., 2012, 13 (12): 789–803. DOI: 10.1038/nrm3474
|
| [21] |
Carroll C W, Silva M C C, Godek K M, et al. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat. Cell Biol., 2009, 11 (7): 896–902. DOI: 10.1038/ncb1899
|
| [22] |
Hinshaw S M, Harrison S C. An Iml3-Chl4 heterodimer links the core centromere to factors required for accurate chromosome segregation. Cell Rep., 2013, 5 (1): 29–36. DOI: 10.1016/j.celrep.2013.08.036
|
| [23] |
McKinley K L, Sekulic N, Guo L Y, et al. The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol. Cell, 2015, 60 (6): 886–898. DOI: 10.1016/j.molcel.2015.10.027
|
| [24] |
Carroll C W, Milks K J, Straight A F. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J. Cell Biol., 2010, 189 (7): 1143–1155. DOI: 10.1083/jcb.201001013
|
| [25] |
Guse A, Carroll C W, Moree B, et al. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature, 2011, 477 (7364): 354–358. DOI: 10.1038/nature10379
|
| [26] |
Tachiwana H, Kagawa W, Shiga T, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature, 2011, 476 (7359): 232–235. DOI: 10.1038/nature10258
|
| [27] |
Fang J N, Liu Y T, Wei Y, et al. Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev., 2015, 29 (10): 1058–1073. DOI: 10.1101/gad.259432.115
|
| [28] |
Tian T, Li X R, Liu Y Y, et al. Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res., 2018, 28 (3): 374–378. DOI: 10.1038/cr.2018.13
|
| [29] |
Chittori S, Hong J, Saunders H, et al. Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science, 2018, 359 (6373): 339–343. DOI: 10.1126/science.aar2781
|
| [30] |
Pentakota S, Zhou K, Smith C, et al. Decoding the centromeric nucleosome through CENP-N. eLife, 2017, 6: e33442. DOI: 10.7554/eLife.33442
|
| [31] |
Yan K, Yang J, Zhang Z, et al. Structure of the inner kinetochore CCAN complex assembled onto a centromeric nucleosome. Nature, 2019, 574 (7777): 278–282. DOI: 10.1038/s41586-019-1609-1
|
| [32] |
Tian T, Chen L, Dou Z, et al. Structural insights into human CCAN complex assembled onto DNA. Cell Discov., 2022, 8 (1): 90. DOI: 10.1038/s41421-022-00439-6
|
| [33] |
Yatskevich S, Muir K W, Bellini D, et al. Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome. Science, 2022, 376 (6595): 844–852. DOI: 10.1126/science.abn3810
|
| [34] |
Pesenti M E, Raisch T, Conti D, et al. Structure of the human inner kinetochore CCAN complex and its significance for human centromere organization. Mol. Cell, 2022, 82 (11): 2113–2131.e8. DOI: 10.1016/j.molcel.2022.04.027
|
| [35] |
Moree B, Meyer C B, Fuller C J, et al. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J. Cell Biol., 2011, 194 (6): 855–871. DOI: 10.1083/jcb.201106079
|
| [36] |
Watanabe R, Hara M, Okumura E I, et al. CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J. Cell Biol., 2019, 218 (218): 4042–4062. DOI: 10.1083/jcb.201907006
|
| [37] |
Liu, R, Dou, Z, Tian, T, et al. Dynamic phosphorylation of CENP-N by CDK1 guides accurate chromosome segregation in mitosis. J. Mol. Cell Biol., 2023: mjad041. DOI: 10.1093/jmcb/mjad041
|
| [38] |
Navarro A P, Cheeseman I M. Dynamic cell cycle-dependent phosphorylation modulates CENP-L-CENP-N centromere recruitment. Mol. Biol. Cell, 2022, 33 (10): ar87. DOI: 10.1091/mbc.E22-06-0239
|
| [39] |
Singh P, Pesenti M E, Maffini S, et al. BUB1 and CENP-U, primed by CDK1, are the main PLK1 kinetochore receptors in mitosis. Mol. Cell, 2021, 81 (1): 67–87.e9. DOI: 10.1016/j.molcel.2020.10.040
|
| [40] |
Chen Q F, Zhang M, Pan X, et al. Bub1 and CENP-U redundantly recruit Plk1 to stabilize kinetochore-microtubule attachments and ensure accurate chromosome segregation. Cell Rep., 2021, 36 (12): 109740. DOI: 10.1016/j.celrep.2021.109740
|
| [41] |
Sedzro D M, Yuan X, Mullen M, et al. Phosphorylation of CENP-R by Aurora B regulates kinetochore-microtubule attachment for accurate chromosome segregation. J. Mol. Cell Biol., 2022, 14 (7): mjac051. DOI: 10.1093/jmcb/mjac051
|
| [42] |
Ariyoshi M, Makino F, Watanabe R, et al. Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C. EMBO J., 2021, 40 (5): e105671. DOI: 10.15252/embj.2020105671
|
| [43] |
Böhm M, Killinger K, Dudziak A, et al. Cdc4 phospho-degrons allow differential regulation of Ame1CENP-U protein stability across the cell cycle. eLife, 2021, 10: e67390. DOI: 10.7554/eLife.67390
|
| [44] |
Westermann S, Cheeseman I M, Anderson S, et al. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol., 2003, 163 (2): 215–222. DOI: 10.1083/jcb.200305100
|
| [45] |
Cheeseman I M, Niessen S, Anderson S, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev., 2004, 18 (18): 2255–2268. DOI: 10.1101/gad.1234104
|
| [46] |
Screpanti E, De Antoni A, Alushin G M, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr. Biol., 2011, 21 (5): 391–398. DOI: 10.1016/j.cub.2010.12.039
|
| [47] |
Przewloka M R, Venkei Z, Bolanos-Garcia V M, et al. CENP-C is a structural platform for kinetochore assembly. Curr. Biol., 2011, 21 (5): 399–405. DOI: 10.1016/j.cub.2011.02.005
|
| [48] |
Petrovic A, Keller J, Liu Y, et al. Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell, 2016, 167 (4): 1028–1040.e15. DOI: 10.1016/j.cell.2016.10.005
|
| [49] |
Gascoigne K E, Takeuchi K, Suzuki A, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell, 2011, 145 (3): 410–422. DOI: 10.1016/j.cell.2011.03.031
|
| [50] |
Schleiffer A, Maier M, Litos G, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat. Cell Biol., 2012, 14 (6): 604–613. DOI: 10.1038/ncb2493
|
| [51] |
Gascoigne K E, Cheeseman I M. CDK-dependent phosphorylation and nuclear exclusion coordinately control kinetochore assembly state. J. Cell Biol., 2013, 201 (1): 23–32. DOI: 10.1083/jcb.201301006
|
| [52] |
Nishino T, Rago F, Hori T, et al. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J., 2013, 32 (3): 424–436. DOI: 10.1038/emboj.2012.348
|
| [53] |
Huis In 't Veld P J, Jeganathan S, Petrovic A, et al. Molecular basis of outer kinetochore assembly on CENP-T. eLife, 2016, 5: e21007. DOI: 10.7554/eLife.21007
|
| [54] |
Yang Y, Wu F, Ward T, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem., 2008, 283 (39): 26726–26736. DOI: 10.1074/jbc.M804207200
|
| [55] |
Dimitrova Y N, Jenni S, Valverde R, et al. Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell, 2016, 167 (4): 1014–1027.e12. DOI: 10.1016/j.cell.2016.10.011
|
| [56] |
Zhou X, Zheng F, Wang C L, et al. Phosphorylation of CENP-C by Aurora B facilitates kinetochore attachment error correction in mitosis. Proc. Natl. Acad. Sci. U.S.A., 2017, 114 (50): E10667–E10676. DOI: 10.1073/pnas.1710506114
|
| [57] |
Hara M, Ariyoshi M, Okumura E I, et al. Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat. Cell Biol., 2018, 20 (12): 1378–1388. DOI: 10.1038/s41556-018-0230-0
|
| [58] |
Foley E A, Kapoor T M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol., 2013, 14 (1): 25–37. DOI: 10.1038/nrm3494
|
| [59] |
Nijenhuis W, von Castelmur E, Littler D, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J. Cell Biol., 2013, 201 (2): 217–231. DOI: 10.1083/jcb.201210033
|
| [60] |
Dou Z, Liu X, Wang W W, et al. Dynamic localization of Mps1 kinase to kinetochores is essential for accurate spindle microtubule attachment. Proc. Natl. Acad. Sci. U.S.A., 2015, 112 (33): E4546–E4555. DOI: 10.1073/pnas.1508791112
|
| [61] |
Hiruma Y, Sacristan C, Pachis S T, et al. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science, 2015, 348 (6240): 1264–1267. DOI: 10.1126/science.aaa4055
|
| [62] |
Ji Z, Gao H, Yu H. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science, 2015, 348 (6240): 1260–1264. DOI: 10.1126/science.aaa4029
|
| [63] |
Primorac I, Weir J R, Chiroli E, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife, 2013, 2: e01030. DOI: 10.7554/eLife.01030
|
| [64] |
Ji Z, Gao H, Jia L, et al. A sequential multitarget Mps1 phosphorylation cascade promotes spindle checkpoint signaling. eLife, 2017, 6: e22513. DOI: 10.7554/eLife.22513
|
| [65] |
Rodriguez-Rodriguez J A, Lewis C, McKinley K L, et al. Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr. Biol., 2018, 28 (21): 3422–3429.e5. DOI: 10.1016/j.cub.2018.10.006
|
| [66] |
Alfonso-Pérez T, Hayward D, Holder J, et al. MAD1-dependent recruitment of CDK1-CCNB1 to kinetochores promotes spindle checkpoint signaling. J. Cell Biol., 2019, 218 (4): 1108–1117. DOI: 10.1083/jcb.201808015
|
| [67] |
Allan L A, Reis M C, Ciossani G, et al. Cyclin B1 scaffolds MAD1 at the kinetochore corona to activate the mitotic checkpoint. EMBO J., 2020, 39 (12): e103180. DOI: 10.15252/embj.2019103180
|
| [68] |
Jackman M, Marcozzi C, Barbiero M, et al. Cyclin B1-Cdk1 facilitates MAD1 release from the nuclear pore to ensure a robust spindle checkpoint. J. Cell Biol., 2020, 219 (6): e201907082. DOI: 10.1083/jcb.201907082
|
| [69] |
Liu S K, Yuan X, Cui P, et al. Mad2 promotes Cyclin B2 recruitment to the kinetochore for guiding accurate mitotic checkpoint. EMBO Rep., 2022, 23 (6): e54171. DOI: 10.15252/embr.202154171
|
| [70] |
Liu X, Liu X, Wang H W, et al. Phase separation drives decision making in cell division. J Biol. Chem., 2020, 295 (39): 13419–13431. DOI: 10.1074/jbc.REV120.011746
|
| [71] |
Xia P, Liu X, Wu B, et al. Superresolution imaging reveals structural features of EB1 in microtubule plus-end tracking. Mol. Biol. Cell, 2014, 25 (25): 4166–4173. DOI: 10.1091/mbc.E14-06-1133
|
| [72] |
Huang Y J, Lin L, Liu X, et al. BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules. Cell Res., 2019, 29 (7): 562–578. DOI: 10.1038/s41422-019-0178-z
|
| [73] |
Song X Y, Yang F R, Liu X, et al. Dynamic crotonylation of EB1 by TIP60 ensures accurate spindle positioning in mitosis. Nat. Chem. Biol., 2021, 17 (12): 1314–1323. DOI: 10.1038/s41589-021-00875-7
|
| [74] |
Song X Y, Yang F R, Yang T T, et al. Phase separation of EB1 guides microtubule plus-end dynamics. Nat. Cell Biol., 2023, 25 (1): 79–91. DOI: 10.1038/s41556-022-01033-4
|
| [75] |
Meier S M, Farcas A M, Kumar A, et al. Multivalency ensures persistence of a +TIP body at specialized microtubule ends. Nat. Cell Biol., 2023, 25 (1): 56–67. DOI: 10.1038/s41556-022-01035-2
|
| [76] |
Maan R, Reese L, Volkov V A, et al. Multivalent interactions facilitate motor-dependent protein accumulation at growing microtubule plus-ends. Nat. Cell Biol., 2023, 25 (1): 68–78. DOI: 10.1038/s41556-022-01037-0
|
| [77] |
Zhang S W, Zou S T, Yin D Y, et al. USP14-regulated allostery of the human proteasome by time-resolved cryo-EM. Nature, 2022, 605 (7910): 567–574. DOI: 10.1038/s41586-022-04671-8
|
| [78] |
Ly P, Brunner S F, Shoshani O, et al. Chromosome segregation errors generate a diverse spectrum of simple and complex genomic rearrangements. Nat. Genet., 2019, 51 (4): 705–715. DOI: 10.1038/s41588-019-0360-8
|
| [79] |
Yao X B, Smolka A J. Gastric parietal cell physiology and Helicobacter pylori-induced disease. Gastroenterology, 2019, 156 (8): 2158–2173. DOI: 10.1053/j.gastro.2019.02.036
|

